Remove Replicate Sampling With Tens of Thousands of Cells Precisely Measured in One Run
The critical component for successful results lies in the prepared sample, be it cultured cells, blood samples, fresh or frozen tissue, or paraffin-embedded sections. For any of these that require cell suspensions or isolated nuclei, accurate and reliable counting to determine the suspensions’ concentration is vital. But significant challenges currently stand in the way of getting it.
The main challenge that has plagued experiments for decades is knowing what the real concentration is. The variability between counting runs of the same sample is typically high, so the number of replicates needed to get your mean concentration close enough to the true mean concentration ends up costing you more time and sample than it’s worth.
One of the biggest causes for this variability is the number of cells counted. When a small number of cells from the sample and it is assumed that it is going to represent the entire sample as a whole, significantly different values are recorded each time. A hemocytometer is a good example of a method that faces this challenge, as it relies on the manual counting of a small number of cells pipetted onto a grid. More modern solutions like image-based cell counters have taken the manual counter out of the technician’s hands, though they still extrapolate the total cell concentration from a few hundred cells. So while some of this challenge has been tackled insofar as the removal of the fallibility of the human eye for counting, image-based counting still has trouble from higher variability between replicates.
A second critical cause for variability between replicates is the proper identification of real cells versus debris. When using a dye to distinguish between the two, the assumption that the dye will properly stain every real cell and not stain debris means that if 10% of the cells are not stained properly, you will by definition insert 10% variability by default even if the counting is perfect. This is especially relevant for blood samples and homogenized tissue samples, as there will be a decent amount of debris and will make reliably counting those samples more difficult. A hemocytometer and most image-based counting systems rely on such dyes, usually Trypan blue, and are therefore susceptible to this.
Because of the low numbers of cells counted and the reliance on dyes for accurate delineation of real cells, it is often accepted that a sample needs to be counted at least three times in order to get something moderately close to the true concentration. This is a waste of precious sample, time, and reagents. An ideal solution would be a device that could solve both these problems and give you a concentration you could rely on without the need for many replicates and does not rely on a dye to find the real cells.
The Moxi Z, with its proprietary coupling of Coulter principle-based cell counting and microfluidics, is the instrument that solves these hurdles. Because the technology relies on detecting changes in electrical current as a particle passes through the aperture, it is able to detect changes down to the microsecond. This allows for the measurement of tens of thousands of cells being pushed through the aperture. A physics-based counting system will never have to rely on visual counting of the cells, which also eliminates the need for a dye to distinguish real cells from debris. Because the cells are sized as they pass through, it is very easy to isolate real cells, which are much larger, from debris, which is much smaller. This gives reliable results within 5% of the true mean on the first run, even in samples with higher debris content like blood and tissue suspensions. Also, because it relies on a simple electrical-based method and not a high-quality camera and AI-based cell detection system, the price point of the Moxi Z is low.
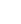
This can be seen with some real-world user data for counting isolated nuclei:
3μL of original sample diluted 20x in PBS gave almost 60,000 nuclei counts from one 10 second run in the microfluidics cassette. With the added ability to gate on specifically nuclei or cells of interest (nuclei = 5-20μm in diameter), it is a unique way to ensure that debris is quickly and simply removed from the counting and concentration calculations (see events in the shaded region above).


-2.png)